7.5. Decision Trees and Random Forest¶
7.5.1. Decision Trees¶
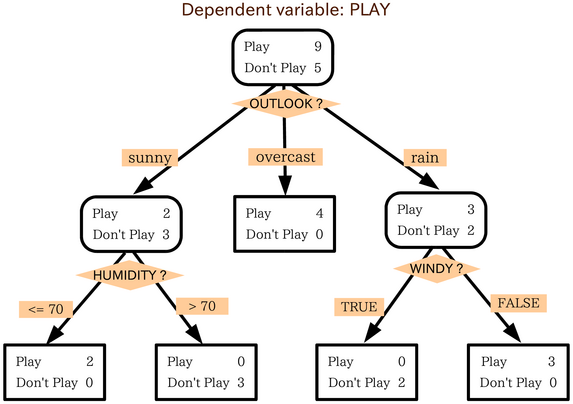 (Image by Wikipedia)
(Image by Wikipedia)
Decision Trees (DTs) are a non-parametric supervised learning method used for classification and regression.
The goal is to create a model that predicts the value of a target variable by learning simple decision rules inferred from the data
features.
A tree can be seen as a piecewise constant approximation.
The deeper the tree, the more complex the decision rules and the fitter the model.
7.5.1.1. Advantages:¶
Simple to understand and to interpret. Trees can be visualised.
Requires little data preparation.
The cost of using the tree (i.e., predicting data) is logarithmic in the number of data points used to train the tree.
Total cost over the entire trees is \(O(n_{features}n_{samples}^2log(n_{samples}))\)Able to handle both numerical and categorical data (However, see disadvatange …).
Uses a white box model rather than a black box model such as neural network. (See figure above)
7.5.1.2. Disadvantages:¶
Decision-tree learners can create over-complex trees that do not generalise the data well. This is called overfitting. Mechanisms such as pruning, setting the minimum number of samples required at a leaf node or setting the maximum depth of the tree are necessary to avoid this problem.
Decision trees can be unstable because small variations in the data might result in a completely different tree being generated. This problem is mitigated by using decision trees within an ensemble.
Predictions of decision trees are neither smooth nor continuous.
Practical decision-tree learning algorithms cannot guarantee to return the globally optimal decision tree.
7.5.1.3. Mathematical formulation¶
7.5.1.4. Classification¶
DecisionTreeClassifier is a class capable of performing multi-class classification on a dataset.
DecisionTreeClassifier takes two arrays as input : an array X, sparse or dense, of shape (n_samples, n_features) holding the
training samples, and an array Y of integer values, shape (n_samples,), holding the class labels for the training samples.
sklearn.tree.DecisionTreeClassifier(*, criterion=’gini’, splitter=’best’, max_depth=None, min_samples_split=2, min_samples_leaf=1, min_weight_fraction_leaf=0.0, max_features=None, random_state=None, max_leaf_nodes=None, min_impurity_decrease=0.0, class_weight=None, ccp_alpha=0.0) details
Splitter:
The splitter is used to decide which feature and which threshold is used.
Using best, the model if taking the feature with the highest importance
Using random, the model if taking the feature randomly but with the same distribution
random_state’s value may be:
None (default)
Use the global random state instance from numpy.random.
Calling the function multiple times will reuse the same instance, and will produce different results.
An integer
Use a new random number generator seeded by the given integer.
Using an int will produce the same results across different calls.
However, it may be worthwhile checking that your results are stable
across a number of different distinct random seeds. Popular integer random seeds are 0 and 42.
%config InlineBackend.figure_formats = ['svg']
from matplotlib import pyplot as plt
from sklearn.datasets import load_iris
from sklearn import tree
# Prepare the data data
iris = load_iris()
X, y = iris.data, iris.target
# Fit the classifier with default hyper-parameters
clf = tree.DecisionTreeClassifier()
clf = clf.fit(X, y)
The data set contains 3 classes of 50 instances each, where each class refers to a type of iris plant. One class is linearly
separable from the other 2; the latter are NOT linearly separable from each other.
Attribute Information:
sepal length in cm
sepal width in cm
petal length in cm
petal width in cm
class:
Iris Setosa
Iris Versicolour
Iris Virginica
sklearn.tree.plot_tree(decision_tree, *, max_depth=None, feature_names=None, class_names=None, label=’all’, filled=False, impurity=True, node_ids=False, proportion=False, rounded=False, precision=3, ax=None, fontsize=None) details
fig = plt.figure(figsize=(12,10))
_ = tree.plot_tree(clf, max_depth=None,
feature_names=iris.feature_names,
class_names=iris.target_names,
filled=True)
text_representation = tree.export_text(clf)
print(text_representation)
7.5.2. Random Forests¶
 (Image by Wikipedia)
(Image by Wikipedia)
Random forests or random decision forests are an ensemble learning method for classification, regression and other tasks that operates by constructing a multitude of decision trees at training time.
7.5.2.1. Advantages:¶
Random Forest is based on the bagging algorithm and uses Ensemble Learning technique. It creates as many trees on the subset of the data and combines the output of all the trees. In this way it reduces overfitting problem in decision trees and also reduces the variance and therefore improves the accuracy.
No feature scaling required: No feature scaling (standardization and normalization) required in case of Random Forest as it uses rule based approach instead of distance calculation.
Handles non-linear parameters efficiently: Non linear parameters don’t affect the performance of a Random Forest unlike curve based algorithms. So, if there is high non-linearity between the independent variables, Random Forest may outperform as compared to other curve based algorithms.
Random Forest is usually robust to outliers and can handle them automatically.
Random Forest algorithm is very stable. Even if a new data point is introduced in the dataset, the overall algorithm is not affected much since the new data may impact one tree, but it is very hard for it to impact all the trees.
7.5.2.2. Disadvantages:¶
Complexity: Random Forest creates a lot of trees (unlike only one tree in case of decision tree) and combines their outputs. By default, it creates 100 trees in Python sklearn library. To do so, this algorithm requires much more computational power and resources. On the other hand decision tree is simple and does not require so much computational resources.
In random forests, each tree in the ensemble is built from a sample drawn with replacement (i.e., a bootstrap sample) from the training set.
class sklearn.ensemble.RandomForestClassifier(n_estimators=100, *, criterion=’gini’, max_depth=None, min_samples_split=2, min_samples_leaf=1, min_weight_fraction_leaf=0.0, max_features=’auto’, max_leaf_nodes=None, min_impurity_decrease=0.0, bootstrap=True, oob_score=False, n_jobs=None, random_state=None, verbose=0, warm_start=False, class_weight=None, ccp_alpha=0.0, max_samples=None) [details](https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestClassifier.html#sklearn. ensemble.RandomForestClassifier)
# Import train_test_split function
from sklearn.model_selection import train_test_split
# Creating a DataFrame of given iris dataset.
import pandas as pd
data=pd.DataFrame({
'sepal length':iris.data[:,0],
'sepal width':iris.data[:,1],
'petal length':iris.data[:,2],
'petal width':iris.data[:,3],
'species':iris.target
})
# data.head()
# Split dataset into features and labels
X=data[['petal length', 'petal width','sepal length']] # Removed feature "sepal length"
y=data['species']
# Split dataset into training set and test set
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.70, random_state=5) # 70% training and 30% test
#Import Random Forest Model
from sklearn.ensemble import RandomForestClassifier
#Create a Gaussian Classifier
clf=RandomForestClassifier(n_estimators=100)
#Train the model using the training sets y_pred=clf.predict(X_test)
clf.fit(X_train,y_train)
y_pred=clf.predict(X_test)
#Import scikit-learn metrics module for accuracy calculation
from sklearn import metrics
# Model Accuracy, how often is the classifier correct
print("Accuracy:",metrics.accuracy_score(y_test, y_pred))
7.5.2.3. Extensions¶